HPC-AI - Introduction to machine learning - Final project
HPC-AI - Introduction to machine learning - Final project
Predicting the function of a protein is one core challenge in biology. Protein function is typically inferred using sequence comparison with proteins whose function is known. This is problematic in many ways. In particular, proteins can have very different sequences, and yet share the same function. Machine learning approaches are thus more and more investigated to go beyond sequence similarity when annotating protein functions in new organism.
This project is about investigating machine learning strategies to infer whether a protein is involved in a secretion system.
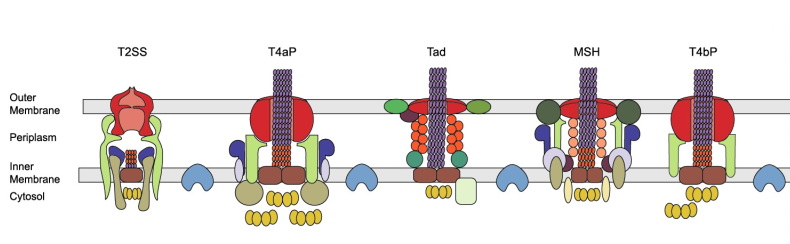
Secretion systems are complexes of proteins enabling a number of essential functions for prokaryotic organisms (bacteria and archaea) such as acquiring nutrients or invading host cells. There are currently 12 types of secretion systems known, but we have very strong evidence to believe there are more secretion systems types: annotating the entier set of genomes with those 12 types of secretion systems shows that some bacteria don’t have any. Because of the importance of secretion systems to interact with the environment and with each other, as well as the difficulty of finding novel secretion systems (12 years of research in an experimental lab for the last type of secretion system found), we believe that there are entire types of secretion systems yet to discover.
The dataset
The dataset can be downloaded here
We downloaded the whole set of complete prokaryotic genomes in 2021 (30,000 genomes in total). We then annotated each protein of each genome as being part of a protein family known to be involved in secretion systems (complexes of proteins that allow the bacteria to secrete a number of elements, including toxins). The annotation of each protein is done through sequence comparison between the protein’s sequence and an “average” sequence of a protein family.
The goal is to predict whether a protein is part of a secretion system.
The data handed in to you is:
The folder
partial_dataset_traincontains the training data at your disposition:protein_sequences.csvcontains two columns. The first one corresponds to a unique identifier for the protein of the formGCA_000474035.1_ASM47403v1.AGX32190.1(organism_ref.protein_ref). Then follows the protein sequence.features.csvcontains a series of features extracted from the protein sequence (amino acid proportion, di-amino acid proportion, entropy, bio physical proporties, deep learning embedding “word 2 vec” approache, etc).labels.csvcontains the information of whether the protein is part of one of the secretion system types we could annotate.complete_labels.csvcontains not only whether the protein is part of one of the secretion system types annotated, but also which of these systems it is part of. Some secretion system types were purposedly ignored in the labels handed to you in the training data.
- The folder
partial_dataset_validcontains the features extracted, the protein sequence, and the labels. - The folder
partial_dataset_testcontains the feature extracted and the protein sequence.
PHASE 1 Preliminary analysis of the dataset and analysis plan
Due date: December 7th, 2022
Send the report (in english) to: nelle.varoquaux@cnrs.fr
The first part of this project is to perform a preliminary analysis of the dataset. You will do this by exploring the labels.csv and complete_labels.csv of the training data.
- How many proteins are in this dataset?
- What is the proportion of positive labels in the dataset? What is the proportion of negative labels? What metric(s) would be appropriate to validate the machine learning pipeline? Justify each of the metrics used.
- Now, looking at the complete list of labels, what is the number of elements in each category? What is the proportion of proteins labeled in two categories?
- Identify and describe three challenges when working on this machine learning problem.
- Devise (on paper) a cross-validation strategy that would enable to check that the machine learning pipeline would extend to secretion systems not annotated in our dataset. You can write a pseudo-algorithm describing the cross validation strategy, or draw a schema explain your thought. Explain how this cross validation strategy is a good way to check for generalization in this setting.
PHASE 2. Predicting protein function from it’s sequence
Due date: January, 18th, 2023 Send the report to: nelle.varoquaux@cnrs.fr
More information will be uploaded on December 8th.
The second part of the project is to implement a strategy to solve the problem at hand. Note that the goal is not necessarily to obtain the best prediction on the test data, but to compare several strategies to solve one or two challenges identified in PHASE1. Some of the difficulties of this particular supervised learning task are:
- The dataset is heavily imbalanced: there are very few positive labels in the training data. This can cause problems both in the learning phase and the validation phase. During the learning phase, algorithms need to be appropriate for imbalanced data or combined with specific strategies to handle imbalanced data. During the validation phase, (1) cross validation needs to be done such that there are positive and negative classes in each training/validation set; (2) metrics need to be appropriate for imbalanced classes (e.g., balanced accuracy, F1-scores, ROC-AUC, see https://machinelearningmastery.com/tour-of-evaluation-metrics-for-imbalanced-classification/ for more information)
- The dataset is very large. Handling such dataset is challenging, and data reduction is required for most learning algorithms to converge in a reasonable amount of time. A common, and sometimes very reasonable strategy, consists in just throwing away a large chunk of data.
- We only have a limited number known protein families implicated in a small number of secretion systems types. Cross-validation needs to be performed with care in order to generalize well.
- To complement point 3, the goal being to predict proteins part of unknown secretion types, cross-validation and model evaluation needs to be done with care, by considering in the test / validation sets all protein associated to a family in the test / validation set. As such, the information in
complete_labels.csvneeds to be used to group all proteins of the same family during cross-validation (e.g., https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GroupKFold.html). - Both positive and negative classes have a wide variety of profiles. Results may be affected by outliers
- We are in positive and unlabeled supervised setting (e.g., the negative class actually corresponds to a mixture of unknown positive class and negative class). Because there are unknown secretion types, we are not able to annotate them. As such, some positive class may be hidden in proteins labeled negative.
Outline of PHASE 2.
Phase 2 of the project is divided into three parts. First, implement a validation strategy based on feedback and indication in this outline. Second, create a small machine learning benchmark on a small dataset. Third, explore and attempt to solve one of the challenges identified during the first phase. While the first and second part are heavily guided, the third part is more an exploration of several strategies based on your interest (and suggestions).
Implement the following cross-validation strategy. Do not hesitate to reach out with questions on slack or by emails. In order to check that the machine learning pipeline generalizes well to unknown secretion system types, protein families involved need to be grouped in the cross-validation setting. As such, create a cross-validation such that each set contains specific groups of protein families (each protein can be part of one or more protein families, which is indicated in the
complete_labels.csv). The validation and test set both contain proteins labeled positives that are part of protein families not annotated in the training set.Creating a machine learning pipeline as a benchmark
- Create a small learning dataset. There are 1210590 proteins in this dataset. Keep only the first 30,000.
import pandas as pd X = pd.read_csv("features.csv", index_col=True) X = X[:30000] y = pd.read_csv("labels.csv", index_col=True) y = y[:30000] …- Create a machine learning pipeline on the small dataset and compare several models:
- Try several feature processing pipelines, several machine learning algorithms.
- Evaluate the performance of the models on the
partial_dataset_validdataset. - Describe your finding in part 1 of the report.
Choose one of the identified challenges.
Look online for strategies to tackle one of the challenge. Implement (possibly using existing strategies) one or more of the strategy, combining this strategy with the pipelines from the previous step. Do not hesitate to reach out to me for guidance on slack
Here are suggestions based on some of the challenges possibly identified during the first phase of the challenges. If you would like to explore another aspect, don’t hesitate to reach out to me via slack to discuss options.
- Tackling imbalanced data There are many ways to deal with imbalanced data, that are easily implementable.
- Find and implement at least one strategies to tackle the issue of imbalance data.
- Compare the results of your strategie(s) with an identical machine learning pipeline without tackling the issue of imbalance data.
- Learning on large datasets You have at hand a fairly large dataset (1210590 proteins). If you choose to tackle this challenge, there are several strategies are possible to learn on such a large dataset: (1) downsampling randomly the data as done for the first part of this project; (2) downsampling the data in a non random fashion (by selecting samples that are fairly different for example); (3) using incremental algorithms, such as mini-batch or online learning algorithms. Don’t hesitate to look for possible solutions online or come up with your on strategies.
- Explore classification accuracy as a function of number of training examples.
- Choose one or several strategies, and compare the results of your strategie(s) with the results of a randomly subsampling data. Justify the learning algorithms explored based on the size of the data.
- Positive and unlabeled learning
- Investigate one or several strategies for PU learning. Through your investigation, try to (1) find publicly available code that implements one or several strategies to tackle PU-learning (python options exist; (2) address the question of validating PU-learning problems
- Implement or use one or several strategie(s) and compare their performance on the validation data. Contrasts those results with performance of the pipeline by considering the simple case “Positive-Negative learning”
Converge on a model/pipeline to solve the problem at hand, and run the code on the test data. Save the results as CSV file. The predicted labels of the train, validation, and test should be provided with the report.
Report
You will be evaluated on a final report. Your report should contain the following elements:
Your full names
- Part 1: Create a machine learning pipeline and evaluate it.
- A discussion of feature processing. Did you standardize the data, chose alternative representations for some features, discarded other features, and why?
- The cross-validated performance, on the training data, of the algorithms you explored. You are strongly encouraged to explore the space of parameters for each of these algorithms. Briefly explain how you did it. Discuss which algorithms/parameters work best.
- Discuss the performance, on the validation data, of one model of each of the five families. Discuss whether the results match your expectations. A discussion of additional models you have tried, insights you have gained (e.g., “This method works well but is difficult to fit” or “This method is not very accurate but is really fast to train”).
- A discussion of your choice of a final model. What are these models, how did you construct them, why do you expect them to be your best proposals? Include tables or figures as you see fit.
- Part 2: Exploring strategies
- Briefly explain the setting of the challenge you chose to solve and why.
- Explain the strategy or strategies to solve this problem you found. If you have found several, justify the ones you chose to test on this machine learning problem.
- Similarly as for part 1, report performance on validation data and contrast this with the simple pipeline developped part 1. Present and try to discuss any unusual findings you may have (such as strategies expected to work better, but failed)
